

An isosurface of a honeybee from the Biological Imaging Center's MRI microscope, and an isosurface of a jade plant from a clinical MRI machine.
Click on the image for a full-size version.


The heterogeneous and irregular geometries of biological objects provide unique computer modeling and scientific visualization challenges. Because of the nature of these objects, the biological and medical communities have a strong need of effective computational tools to manage the collection, classification, visualization, and simulation of MRI data. Therefore, research in computer graphics methods for interaction with MRI data collected from actual biological organisms is both pushing the theoretical frontiers and is well-grounded in an immediate application environment.


An isosurface of a honeybee from the Biological Imaging Center's
MRI microscope, and an isosurface of a jade plant from a clinical MRI
machine.
Click on the image for a full-size version.
The Center for Graphics and Visualization is closely involved in all the stages of data manipulation. Through the Biological Imaging Center at Caltech, the center has access to an MRI microscope capable of taking data at ten micron resolution, as well as access to clinical MRI machines at the Huntington MRI Center in Pasadena. Researchers in the Science and Technology Center are collaborating closely with the Biological Imaging Center scientists to better monitor the data collection procedures, in order to classify materials which are not normally discernible in MRI. The involvement of computer graphics modeling has allowed a goal-directed data acquisition scheme which can selectively seek out specific materials interesting to the user.
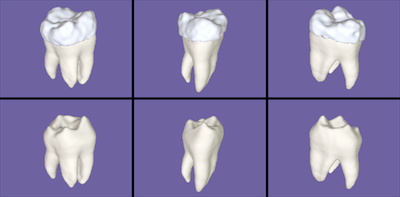
A "clickable map" to 6 separate tooth images: click to see a full-size
image of a tooth. The imaging of water-free solids such as bone and tooth
enamel with MRI is a difficult and very promising research area.
In order to visualize and analyze the data from an MRI volume data set, a technique has been developed which allows voxels to be classified by the amounts of various materials which they contain. This method matches Gaussians and special "mixing" material basis functions in histogram space to best match the imaged data. This has proven to be extremely effective at extracting specific materials from MRI data, used for visualization by "computational staining" and to produce accurate geometric models of the internal structures of biological objects usable in simulations. Details of this work are available in a technical report .
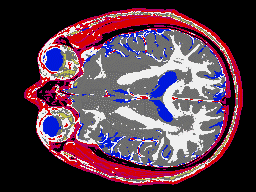
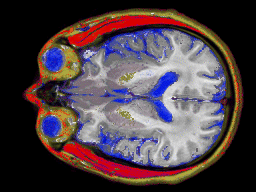
Two images of an MRI ``slice'' through a human brain. The first is
classified with a simple material choice at each pixel, and the second
is calculating mixtures of materials for every pixel for a more
accurate representation.
More classified slice data appears in a movie of axial cross sections of tendons in the hand and one of axial cross section of fat distribution in the hand (both starting from the finger tips).
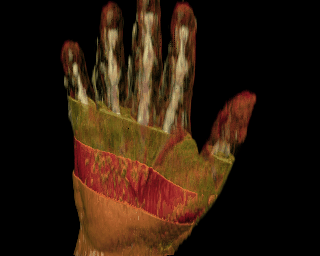
Once the data is classified, it is possible to render it by volume ray-tracing, ray tracing an isosurface, or matching a polygonal representation of an isosurface to the data. One particularly useful method is to deform part of the data based on its classification, thereby allowing the viewer to see the dynamic physical relationships between bones, muscles, and tendons without using invasive techniques.
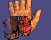



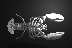
Click on the images to see a hand with the skin ``peeled away'' with a
deformation for a better view of the underlying structures and an isosurface
rendering of a lobster. Also available are two movies of
the hand waving
and
the hand spinning.
This idea of non-invasive biological data collection is useful in studies of in-vivo physiology, anatomy, and developmental biology, as well being a valuable medical visualization tool. The shapes extracted from these techniques can be used in simulations interacting with the generative modeling system as seen in the MRI-imaged lobster data set ``rendered'' in the 1993 SIGGRAPH film Fruit Tracing.


An image of a tadpole from the Biological imaging center MRI microscope
used for developmental biology studies and a frame from a physical simulation
of a peeling banana based on an unpeeled shape extracted from MRI data.
Click on the images to see the full-size versions.